High-Performance Computing (HPC) Cluster Guide¶
BSIC operates a scientific computational resource to help BSIC users to preprocess and analyze neuroimaging data. Due to high load on this resource, we recommend that users postpone their jobs until evening and weekend hours.
Introduction¶
What is it?¶
An HPC cluster is a collection of interconnected computers (nodes) that work together to perform complex computational tasks. The BSIC HPC cluster is designed to provide researchers with the computational power needed to analyze large datasets, such as those generated in neuroimaging studies. The cluster consists of multiple compute nodes, each equipped with powerful processors and large amounts of memory, allowing users to run computationally intensive analyses efficiently. Users can submit their jobs to the cluster's job scheduler, which manages the allocation of resources and ensures that jobs are executed in an orderly manner. The cluster also provides access to a large temporary storage pool, allowing users to store intermediate results and large datasets during their analyses.
Below is a simplified topology of the BSIC HPC cluster.
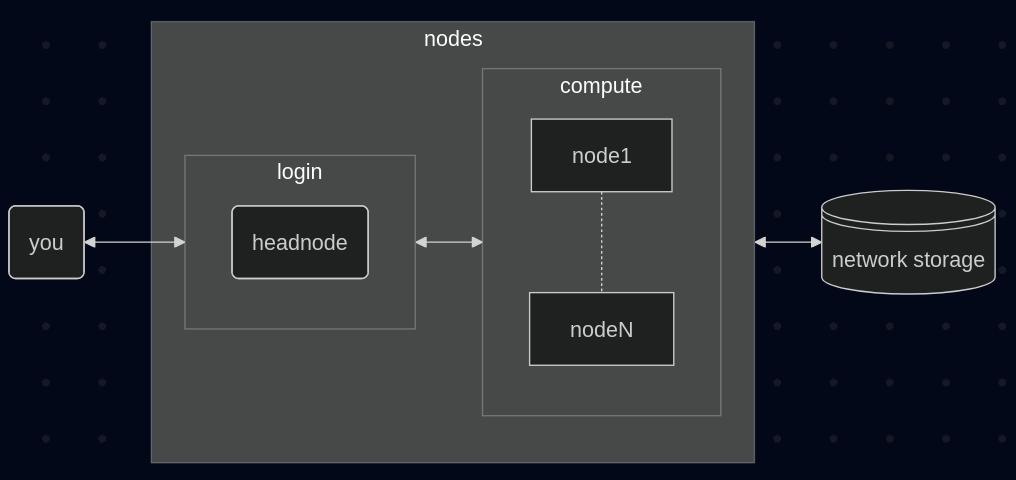
You log into the login node, which is your gateway to the cluster. From there you can submit jobs to the job scheduler (PBS) that will run on the compute nodes. Lastly, nodes have access to a large temporary storage pool.
Why use it?¶
The BSIC HPC cluster provides a powerful computational environment for neuroimaging data analysis. It allows users to run complex analyses on large datasets that would be impractical to run on a personal computer. The cluster's high-performance computing capabilities enable faster processing times, making it possible to analyze data more efficiently and effectively.
How do I get an account?¶
Please see William Burroughs or Peter Kochunov if additional computation capacity is needed, it maybe available through collaborative agreement with the institute for genomic science or off-campus vendors.
Getting Started¶
Logging in¶
You will receive an email with instructions on how to log into the cluster. Please refer to that email for the specific instructions.
Education/Tutorials¶
What is Linux?¶
GNU Linux is a 100% free and open-source operating system that is widely used in scientific computing. It provides a stable, performant, and lightweight environment for running computationally intensive tasks, making it an ideal choice for high-performance computing clusters. BSIC's HPC cluster runs on Linux, so familiarity with Linux is essential for using the cluster effectively.
Basic Linux Commands¶
Navigation & File Management¶
| Task | Command | Notes / Example |
|---|---|---|
| Print working directory | pwd |
|
| List files (detailed) | ls -lh |
human-readable sizes |
| List including hidden | ls -lah |
|
| Change directory | cd /path/to/dir |
cd ~ for home, cd - previous |
| Make directory | mkdir -p /path/to/dir |
-p creates parents |
| Copy file/dir | cp file destcp -r dir dest |
|
| Move/rename | mv src dest |
|
| Remove file | rm file |
|
| Remove directory | rm -rf dir |
use with caution |
| View file start/end | head file / tail file |
-n 50 to set lines; tail -f follow |
| Concatenate files | cat file |
|
| Pager | less file |
navigate large files (q to quit) |
| Disk usage (dir) | du -sh /path |
summary human-readable |
| Disk free (FS) | df -h |
File Permissions & Ownership¶
| Task | Command | Notes |
|---|---|---|
| Show permissions | ls -l |
|
| Change permissions | chmod 640 file |
symbolic: chmod u+rwx,g+rx,o-r |
| Change owner | chown user:group file |
requires permissions (root or sudo) |
Text Processing & Search¶
| Task | Command | Notes / Example |
|---|---|---|
| Search text | grep -n "pattern" file |
-r recursive, -i ignore case |
| Count lines/words/chars | wc -l file |
wc -w words |
| Sort lines | sort file |
sort -k2,2 -n numeric on column 2 |
| Unique lines | uniq |
often used with sort |
| Filter and view | awk '{print $1,$3}' file |
field-based processing |
| Stream editing | sed 's/old/new/g' file |
Compression & Transfer¶
| Task | Command | Notes |
|---|---|---|
| Create tar.gz | tar czf archive.tar.gz /path |
c create, z gzip, f file |
| Extract tar.gz | tar xzf archive.tar.gz |
|
| Compress gzip | gzip file / gunzip file.gz |
|
| scp copy (remote) | scp file user@host:/path |
|
| rsync (efficient) | rsync -avP local/ user@host:/path |
-P shows progress & partials |
| Transfer with port | scp -P 2222 file user@host:/path |
Environment & Modules¶
| Task | Command | Notes |
|---|---|---|
| List env vars | env or printenv |
|
| Set env var (bash) | export VAR=value |
persistent in shell rc files |
| Load software module | module avail / module load python/3.10 |
Common on clusters (Environment Modules or Lmod) |
| List loaded modules | module list |
|
| Unload module | module unload name |
Resource Monitoring & Debugging¶
| Task | Command | Notes |
|---|---|---|
| Show running processes | ps aux | grep myprog |
|
| Interactive process viewer | top or htop |
htop often not installed |
Useful Shortcuts & Tips¶
- Background job:
command &; bring to foreground:fg - Time a command:
/usr/bin/time -v ./program(detailed resource usage) - Edit files:
vim file
What is PBS?¶
Portable Batch System (PBS) is a job scheduling system commonly used on high-performance computing clusters. It allows users to submit computational jobs to a queue, which are then executed on the cluster's compute nodes when resources become available. PBS provides a way to manage and allocate cluster resources efficiently, ensuring that multiple users can run their jobs without conflicts. Users can specify resource requirements (e.g., number of CPUs, memory, runtime) for their jobs, and PBS will handle the scheduling and execution based on these requirements.
PBS Job Examples¶
See the following examples.
Transferring Files¶
There are several ways to transfer files to and from the BSIC HPC cluster.
Using a command-line terminal¶
All Operating Systems (Windows, MacOS, Linux) have built-in command-line
terminals that can be used to transfer files using the scp command. The scp
command allows you to securely copy files between your local machine and the
cluster.
Below are examples of how to use scp to transfer files. Substitute user with
your username and cluster with the hostname or IP address of the cluster.
- To copy a file from your local machine to the cluster:
scp /path/to/local/file user@cluster:/path/to/remote/directory - To copy a file from the cluster to your local machine:
scp user@cluster:/path/to/remote/file /path/to/local/directory - To copy a directory from your local machine to the cluster:
scp -r /path/to/local/directory user@cluster:/path/to/remote/directory - To copy a directory from the cluster to your local machine:
scp -r user@cluster:/path/to/remote/directory /path/to/local/directory
Using a graphical file transfer (GUI) tool¶
FileZilla is a popular graphical file transfer tool that supports the SFTP protocol, which can be used to transfer files to and from the BSIC HPC cluster. To use FileZilla, you will need to configure it with the hostname or IP address of the cluster, your username, and your password. Once connected, you can easily drag and drop files between your local machine and the cluster.
Once you have FileZilla installed, follow these steps to connect to the BSIC HPC cluster:
- Open FileZilla and go to "File" > "Site Manager".
- Click "New Site" and enter a name for the connection (e.g., "BSIC HPC Cluster").
- In the "Host" field, enter the hostname or IP address of the cluster.
- Set the "Port" to 22 (the default for SFTP).
- Set the "Protocol" to "SFTP - SSH File Transfer Protocol".
It is recommended to NOT save your password in FileZilla. Lastly, click "Connect" to establish the connection. You will be prompted to enter your password each time you connect. Once connected, you can navigate the file system of the cluster and transfer files by dragging and dropping them between the local and remote panes.
Software on the Cluster¶
BSIC maintains a large library of scientific software. The software can be categorized into two main groups:
-
Administrator installed software:
$FREEWARE: A collection of commonly used scientific software that is available to all users. This includes software such as MATLAB, FSL, AFNI, FreeSurfer, and many others. This is a variable holding a path to the directory where the software is installed.- System directories (
/usr/bin, etc.): A collection of software that is available to all users. This includes software such as Python, R, and various bioinformatics tools. modulecommand: Some software may be available as modules that can be loaded using themodulecommand. This allows users to easily switch between different versions of software and manage their environment.
-
User installed software:
- User
$HOMEdirectories: Users are welcome to install software in their home directories. However, there is a disk quota and urls are blocked on the cluster, so they may need to download their software on their local machine and then transfer it to the cluster.
- User
When searching for software, it is recommended to check in the following order:
- By using the
modulecommand.- example:
module avail matlab
- example:
- By using the
which,whereis, andlocatecommands.- examples:
which matlabto see if matlab is in your PATH.whereis matlabto see if matlab is installed and where it is located.locate matlabto see if matlab is installed and where it is located.
- examples:
- By checking the
$FREEWAREdirectory.- example:
ls $FREEWAREto see what software is available in the freeware directory.
- example:
If you've gone through these steps and still cannot find the software you need, please contact William Burroughs to see if it can be installed on the cluster.
Containerization¶
Some analysis software may be available as container images. The cluster
supports running containerized software using singularity.
Some containers have been built already and are available by using the
module command. Others may be available in the $FREEWARE directory. If a
software that you need references using docker or any other containerization
software, then you need to build a singularity image from the docker image.
If you need help with building the image, please contact William Burroughs.
Frequently Asked Questions¶
How do I log in?
How do I change my password?
You can use the command passwd.
How can I stay informed about system changes?
William Burroughs maintains a mailing list for all accounts created on the cluster.
Can I run jobs on the login nodes?
Please do not run any jobs on the login node. Running computationally intensive jobs on the login node can cause problems for other users. Processes causing problems on the login node may be terminated without notice.
Disk quota exceeded
Hard quota of 500GB is maintained for home directory. Please see William Burroughs if you need large temporal space for data analyses. Investigators are expected to maintain their own analysis data space. Please see William Burroughs for more details.
Why are my #PBS options being ignored?
Make sure that your #PBS options come before any commands in your shell
script. Once the queuing system encounters a command, it quits looking
for #PBS options, so these options will be ignored.
Where can I find x program?
Is there a backup of my data?
Please note that regular backup of user home space is user's responsibility. BSIC only does monthly backups of the home directory. Back up of project scratch space is responsibility of individual users. Please see William Burroughs on the available options.